Collective Migration during Cancer Progression
Rachel Lee, Postdoctoral Research Associate
with Dr. Stuart Martin (University of Maryland School of Medicine)
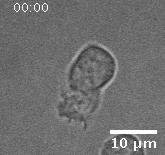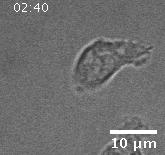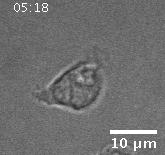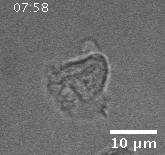
In addition to playing a role in processes such as wound healing and development, collective migration is seen in the progression of diseases such as cancer. As tumor cells become more malignant, they gain the ability to migrate throughout the body; in addition to migrating as individual cells, they have been seen to migrate collectively in vivo and there is increasing evidence that collective behavior plays a role metastsis.
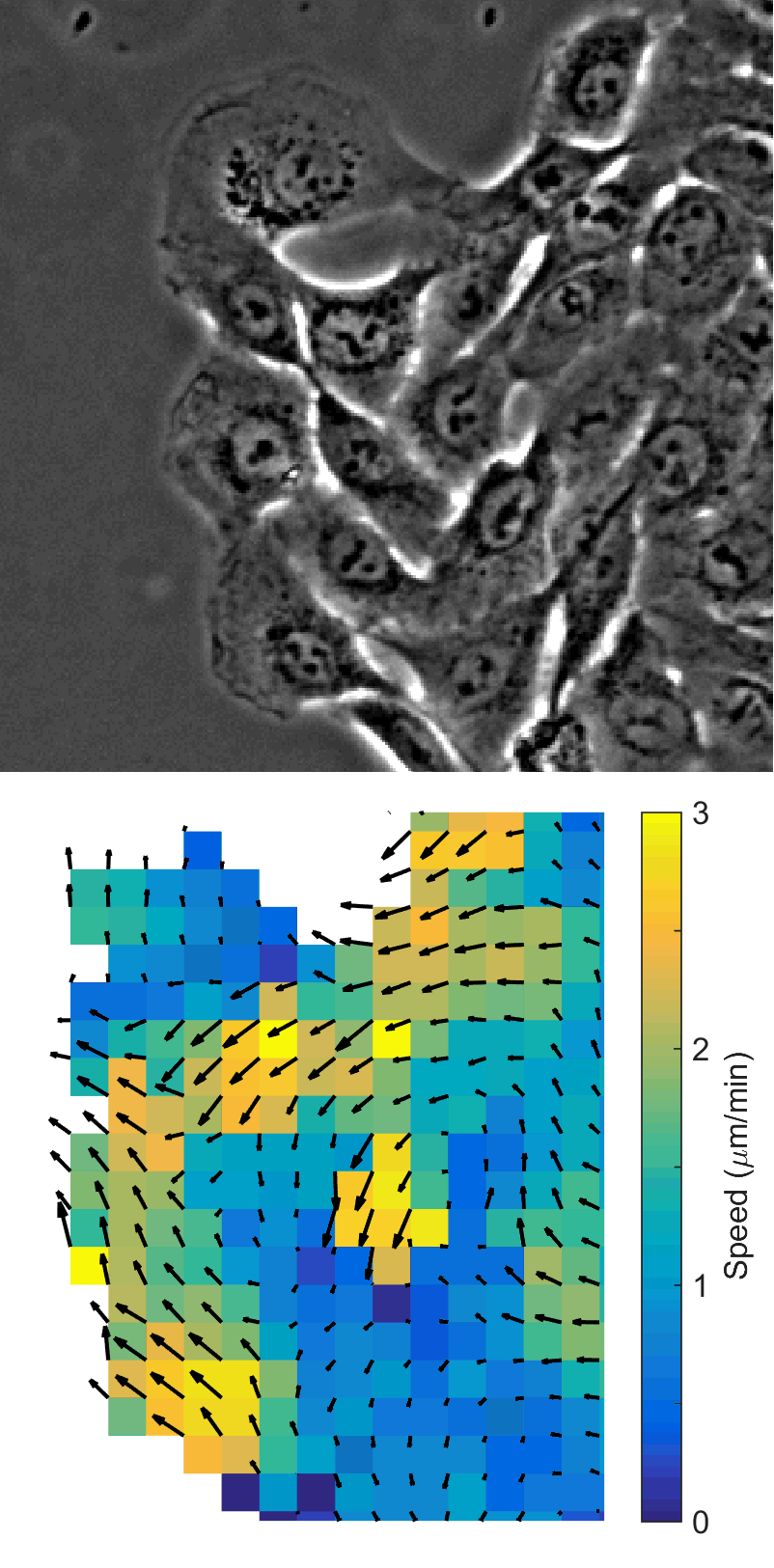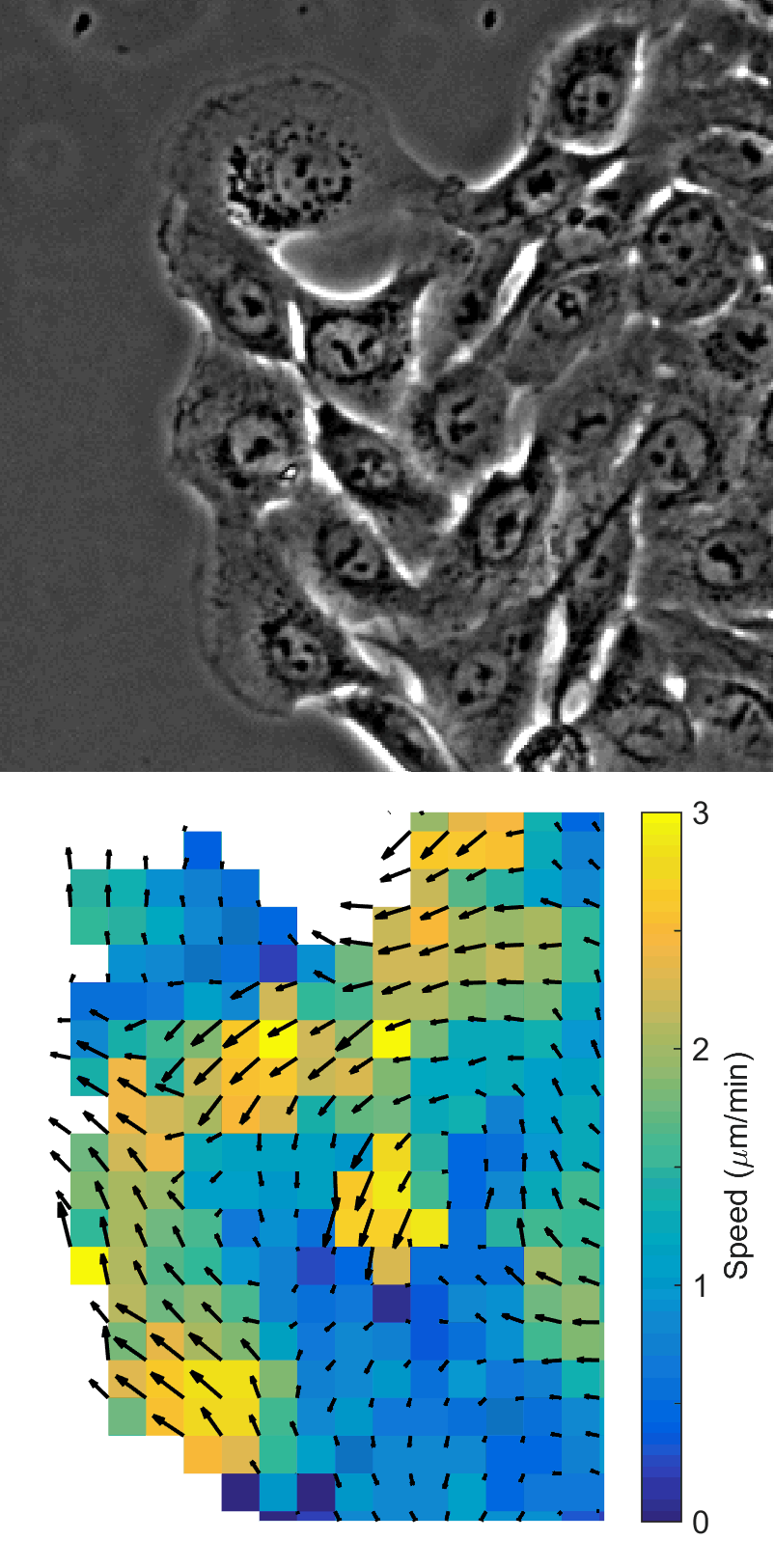
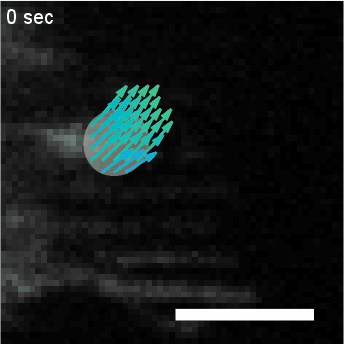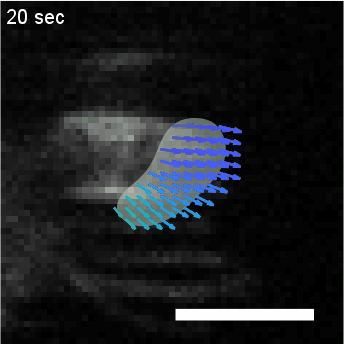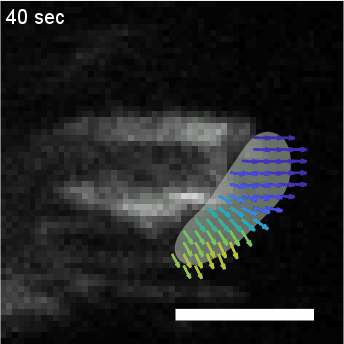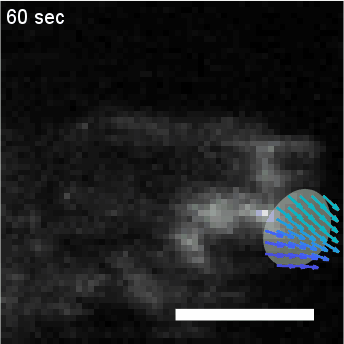
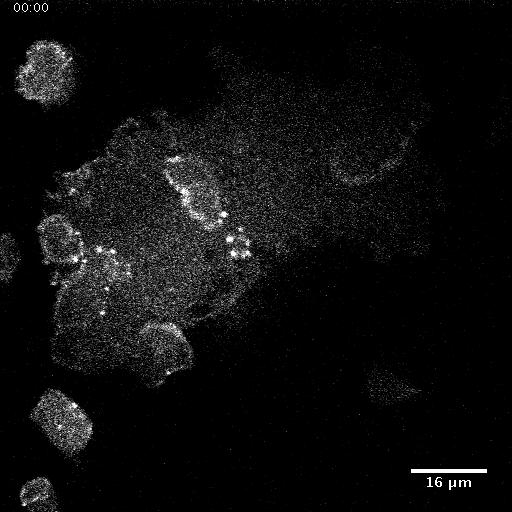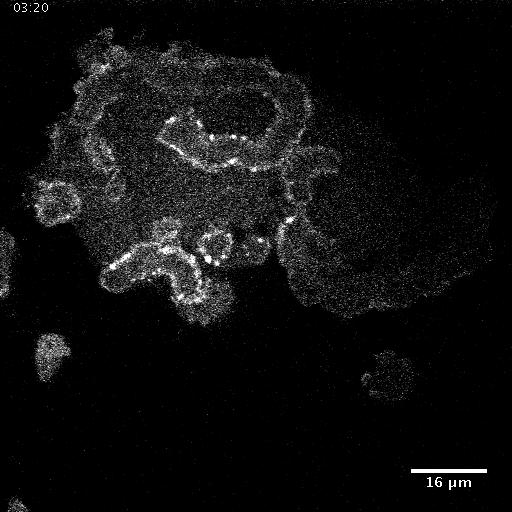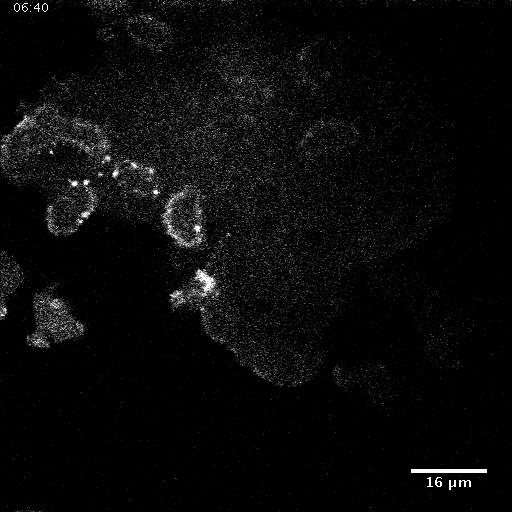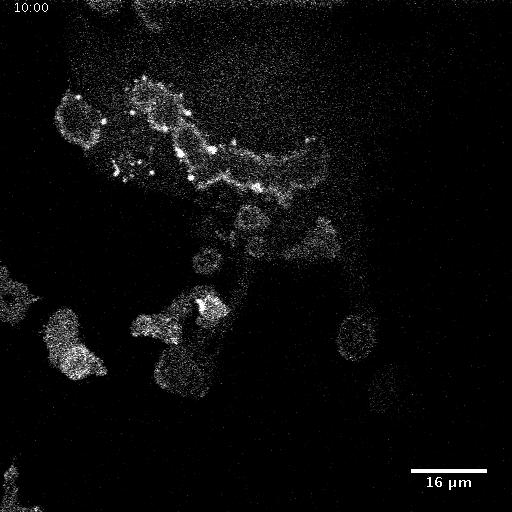
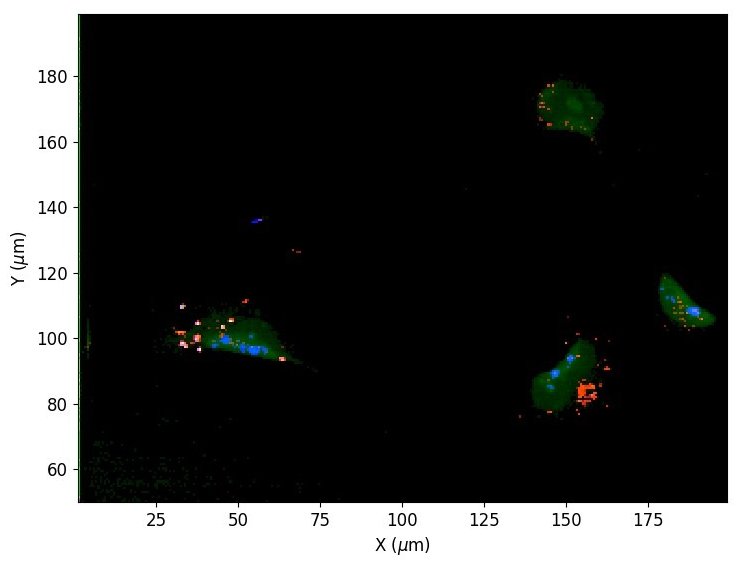
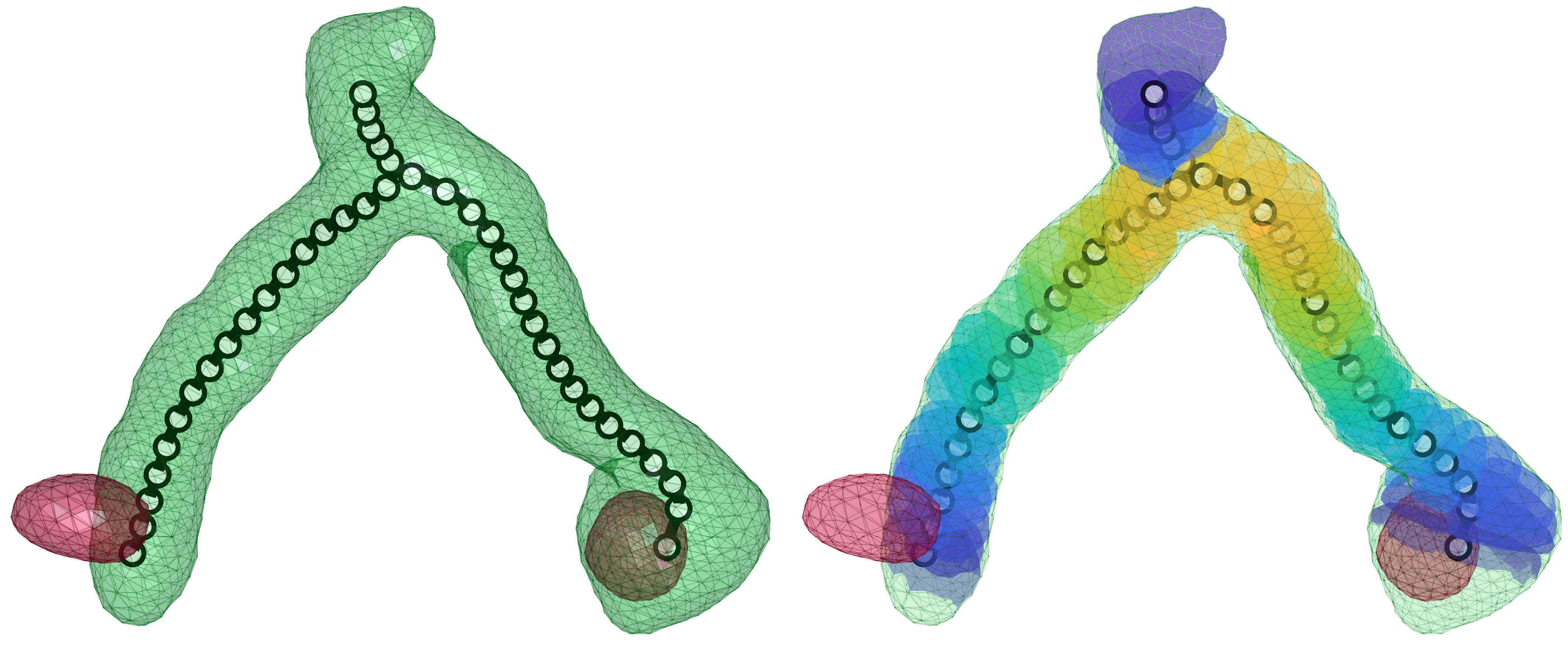
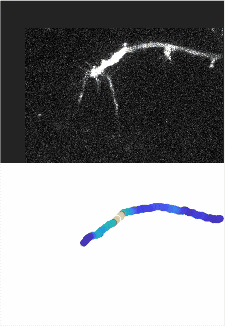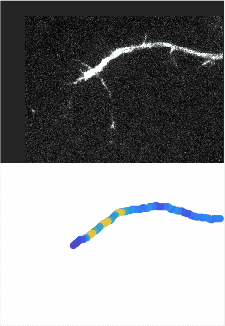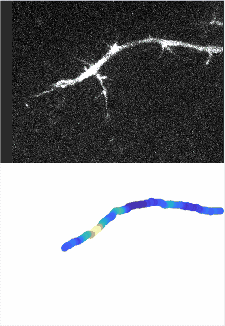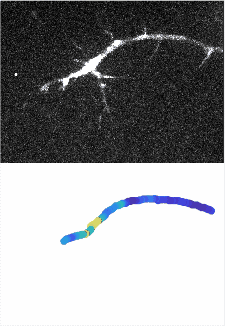
Using automated image analysis techniques we are able to extract information such as velocities from images of migrating human epithelial cells. Inspired by tools developed to study fluid flows and moving grains of sand, we have quantified the motion of migrating epithelial sheets and measured differences in malignant and non-malignant cells. We are currently using these tools to understand how the motion of epithelial cells is regulated and how changes in cell migration are linked to metastatic cancer.
Publications:
R.M. Lee, D.H. Kelley, K.N. Nordstrom, N.T. Ouellette, and W. Losert, New J Phys 2013
R.M. Lee, C.H. Stuelten, C.A. Parent, W. Losert, CSPO 2016
R.M. Lee, H. Yue, W.J. Rappel, W. Losert, Interface 2017
Y. Zhang, X. Guoqing, R.M. Lee, Z. Zhu, J. Wu, S. Liao, G. Zhang, Y. Sun, et. al., Cellular and Molecular Life Sciences 2017
C.H. Stuelten, R.M. Lee, W. Losert, and C.A. Parent, Cellular Signalling 2018
R.M. Lee and W. Losert, Seminars in Cell & Developmental Biology 2019
R.M. Lee, M.I. Vitolo, W. Losert, and S.S. Martin, Scientific Reports 2021